本所夏復國與陳宗岳老師合作提出「海洋病毒分流假說」 研究刊登於全球頂尖期刊《Science Advances》(Impact Factor=14.98)
海洋病毒-細菌間三十年的「恩怨情仇」:病毒分流假說
眾所周知,病原性的細菌與病毒有著毀滅人類族群、社會結構乃至文明發展的黑歷史。鼠疫桿菌(黑死病) 在中世紀奪走了歐洲約半數的人口;冠狀病毒 (COVID-19) 造成了人類歷史上致死人數最多 (645萬;2022年8月前) 的案例。較少為人知的是:占總群聚量95%以上的「非病原性」微生物卻是主導著地球生態系統內物質運作不可或缺的主角。
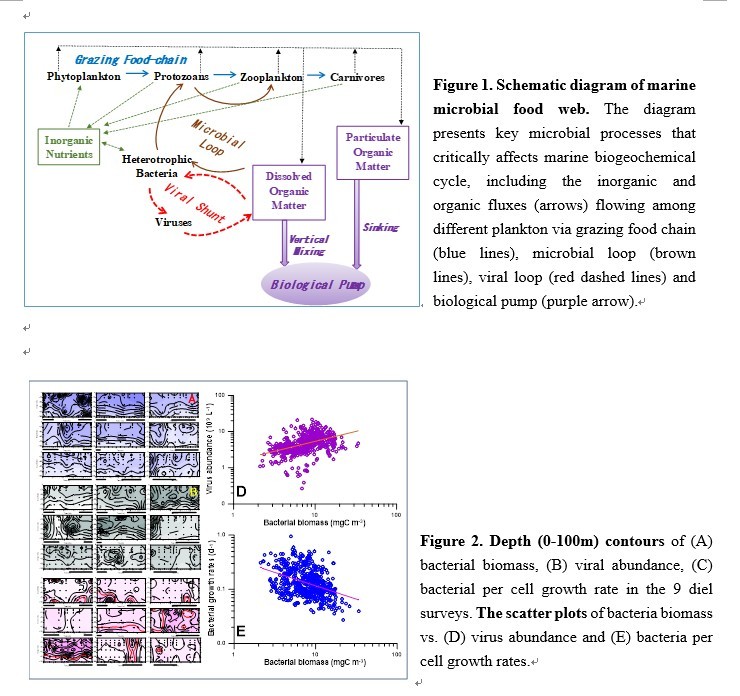
除了 “捕食食物鏈“ (亦即大魚吃小魚,小魚吃蝦米,蝦米吃浮游植物) 外,所有生物生活與死亡後產生的溶解態有機碳 (DOC; 其存量與大氣中的CO2相當,約660 Giga-ton) 只會被異營性細菌吸收用以繁衍。當細菌被原生動物 (鞭/纖毛蟲) 捕食,而原生動物再被浮游動物捕食,透過此一“微生物環“途徑,可以將DOC送回“捕食食物鏈“ (圖1)。
水域環境中約10~50 %細菌的死亡源於病毒感染。“病毒分流“假說 (viral shunt hypothesis;圖1) 認為細菌因病毒感染的數量減少會降低“微生物環“中細菌顆粒碳的向上傳輸;在這個同時,細菌裂解後釋放出的溶菌液 (lysate) 會刺激現存細菌的生長率。然而在過去將近三十年裡,“病毒分流“現象僅局限於實驗室的觀察或數值模式模擬的結果。在自然界中的存在一直是個未解之謎。研究團隊懷疑這可能與野外採樣頻度及研究系統本身特性有關。
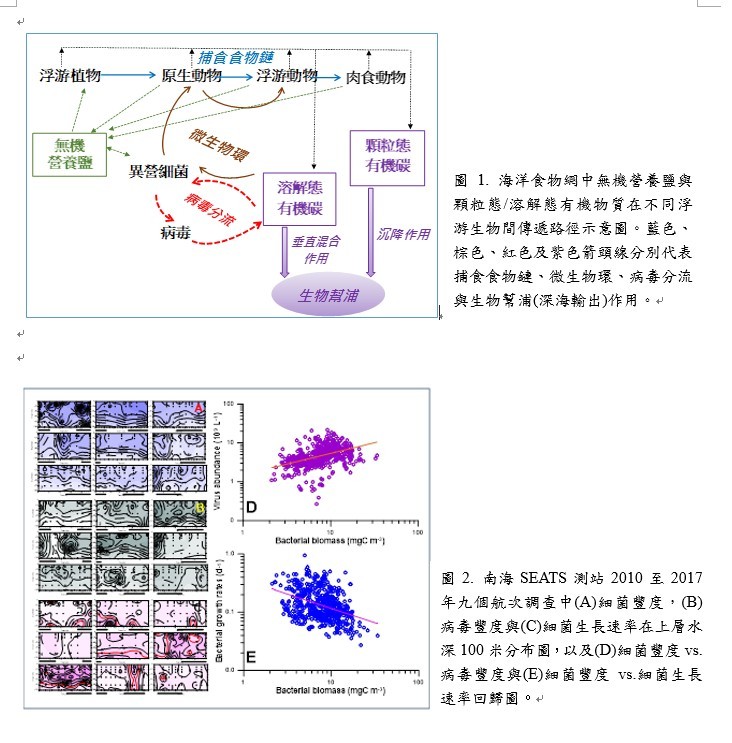
為此,本所陳宗岳副教授與合聘教授中央研究院夏復國研究員偕同臺灣大學團隊,在2010到2017年間,進行並分析了九次南海蹲站調查 (每3小時採樣一次;觀察24~ 48小時) 的數據。結果顯示在時空分布上,細菌豐度(圖2A)與病毒豐度(圖2B)極為相似(圖2D),但二者的豐度皆與細菌生長速率(圖2C)呈負相關(圖2E)。
研究結果有二大重點: (一) 海洋中的浮游細菌長期處於飢餓狀態,稍有”好吃的 (labile)”有機物質的供應,細菌的成長速率反應時間可在數小時之內完成。病毒分流可在數小時內發生並完成而且只能在貧營樣鹽(oligotrophic)的環境下被觀察到;(二) 高溫環境極可能會壓抑病毒感染裂解細菌及其分流作用。研究成果發表於2022年10月12日刊印的《科學進階》(Science Advances)期刊。
參考文獻: Shiah FK, CC Lai, TY Chen, CY Ko & CW Chang. 2022. Viral shunt in tropical oligotrophic ocean. Science Advances. DOI: 10.1126/sciadv.abo2829.
Viral shunt: A 30 years’ unsolved grudge between marine viruses and bacteria.
Pathogenic bacteria and viruses are notorious for destroying the human population and civilization. Yersinia pestis (aka. plague, the black death) took half of the Europeans' life in the Middle Ages, and that COVID-19 has resulted in the highest casualty in human history (6.45 million by August 2022). However, the vast majority (>95%) of these microbes are harmless to us. These non-pathogenic microbes play essential roles in recycling the primary elements that make up all living systems on earth.
The ocean has an inventory of dissolved organic carbon (DOC; ~660 Giga-ton C) roughly equal to that of atmospheric CO2. Most oceanic DOC is derived from autochthonous food web processes. Heterotrophic bacteria are responsible for DOC decomposition and constitute a key component in the “microbial-loop” (Fig. 1), which emphasizes the connections between DOC, bacteria, protozoans, and the traditional grazing (phytoplankton to zooplankton to fishes) food chain.
Viruses are thought responsible for c.a. 10~50% of the total bacterial mortality in surface seawater. The viral shunt hypothesis suggested that the viral-lysis process lessened the transfer of bacterial cells to protozoans; and that the lysate released from the broken cells stimulated the growth of the existing bacteria. This viral shunt hypothesis is crucial for microbial biogeochemistry but its applicability to the real world has not been well documented for almost three decades. We suspected that that the timescale adopted in sampling and system trophic status determine the “visibility” of the viral shunt in the field.
The results from 9 diel surveys (sampling with a 3-hours interval for 24-48 hours; 2010-2017) conducted in the tropical South China Sea indicated that in terms of tempo-spatial distribution, bacterial abundance (Fig. 2A) and virus abundance (Fig. 2B) changed synchronously (Fig. 2D). While bacteria per cell growth rate (Fig. 2C) varied negatively with that of bacteria (and virus too) abundance (Fig. 2E).
Bacteria are in famine status in an oligotrophic ocean. The supply of labile substances such as the lysate from viral lysis process stimulates bacteria to turnover in a time-scale of hours. The difficulty of detecting the viral shunt in mesotrophic/eutrophic systems might due to their abundant allochthonous and autochthonous organics input sources. The differential temperature responses of the viral shunt modes in the upper and lower euphotic zones provides insights into the viral shunt and the consequential impacts on food web processes (i.e. fueling primary production and bacteriovory) in a warming climate. This research has been published in Science Advances on October 12th, 2022.
Reference: Shiah FK, CC Lai, TY Chen, CY Ko & CW Chang*. 2022. Viral shunt in tropical oligotrophic ocean. Science Advances. DOI: 10.1126/sciadv.abo2829. Symbol *, corresponding author.